Iso Distance Ellipsoid
Purpose : Iso Distance Ellipsoid for a specific covariance matrix
> A <- matrix(data = c(1, 2, 3, 2, 1, 2, 3, 2, 1), nrow = 3) > lambda <- diag(3) > diag(lambda) <- eigen(A)$values > eigenvec <- eigen(A)$vectors |
Replicating A using eigen vec
> print(eigenvec %*% lambda %*% t(eigenvec))
[,1] [,2] [,3]
[1,] 1 2 3
[2,] 2 1 2
[3,] 3 2 1
> print(A)
[,1] [,2] [,3]
[1,] 1 2 3
[2,] 2 1 2
[3,] 3 2 1 |
Are the vectors orthogonal ?
> round(t(eigenvec[, 1]) %*% eigenvec[, 2])
[,1]
[1,] 0
> round(t(eigenvec[, 1]) %*% eigenvec[, 3])
[,1]
[1,] 0
> round(t(eigenvec[, 2]) %*% eigenvec[, 3])
[,1]
[1,] 0 |
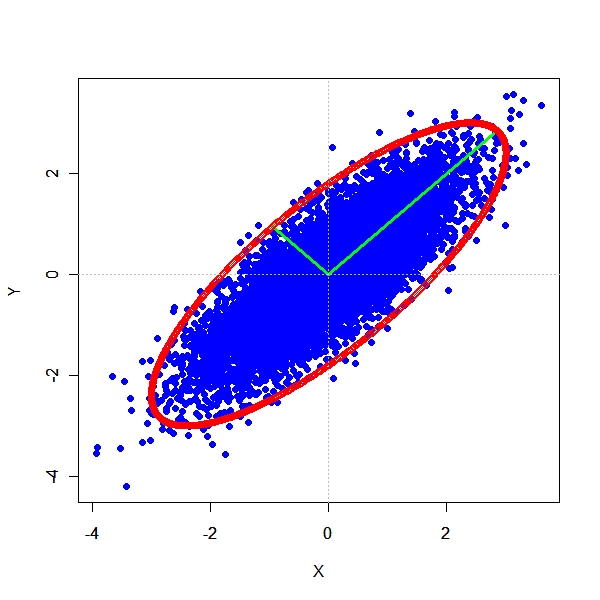
Dependent Bivariate Normal - Cor = 0.8
> TwoDimEllipsoid <- function(sample.mean, sample.cov, Scale) {
+ x1 <- sample.mean[1] - sqrt(sample.cov[1, 1]) * Scale
+ x2 <- sample.mean[1] + sqrt(sample.cov[1, 1]) * Scale
+ y1 <- sample.mean[2] + sqrt(sample.cov[2, 2]) * Scale
+ y2 <- sample.mean[2] - sqrt(sample.cov[2, 2]) * Scale
+ xrange <- c(range(x[, 1])[1] - sample.mean[1], range(x[,
+ 1])[2] + sample.mean[1])
+ yrange <- c(range(x[, 2])[1] - sample.mean[2], range(x[,
+ 2])[2] + sample.mean[2])
+ par(mfrow = c(1, 1))
+ plot(x, pch = 19, col = "blue", xlab = "X", ylab = "Y", main = "",
+ xlim = xrange, ylim = yrange)
+ vecs <- eigen(sample.cov)$vectors
+ evals <- eigen(sample.cov)$values
+ length1 <- sqrt(evals[1])
+ length2 <- sqrt(evals[2])
+ segments(sample.mean[1], sample.mean[2], sample.mean[1] +
+ vecs[1, 1] * length1 * Scale, sample.mean[2] + vecs[2,
+ 1] * length1 * Scale, lwd = 3, col = "green")
+ segments(sample.mean[1], sample.mean[2], sample.mean[1] +
+ vecs[1, 2] * length2 * Scale, sample.mean[2] + vecs[2,
+ 2] * length2 * Scale, lwd = 3, col = "green")
+ par(new = T)
+ angles <- seq(from = 0, to = 2 * pi, by = pi/500)
+ ang.points <- matrix(data = NA, nrow = length(angles), ncol = 2)
+ for (i in seq_along(angles)) {
+ ang.points[i, 1] <- (cos(angles[i])) * sqrt(evals[1]) *
+ Scale
+ ang.points[i, 2] <- (sin(angles[i])) * sqrt(evals[2]) *
+ Scale
+ }
+ ellipsoid <- matrix(data = NA, nrow = n, ncol = 2)
+ ellipsoid <- (ang.points %*% t(vecs))
+ ellipsoid[, 1] <- (ellipsoid[, 1]) + sample.mean[1]
+ ellipsoid[, 2] <- (ellipsoid[, 2]) + sample.mean[2]
+ plot(ellipsoid, xlab = "", ylab = "", xlim = xrange, ylim = yrange,
+ col = "red")
+ abline(v = sample.mean[1], lty = "dotted", col = "grey")
+ abline(h = sample.mean[2], lty = "dotted", col = "grey")
+ }
> n <- 10000
> rho <- 0.8
> sample.mean <- c(0, 0)
> sample.cov <- matrix(data = c(1, rho, rho, 1), nrow = 2, ncol = 2)
> x <- rmnorm(n = n, mean = sample.mean, varcov = sample.cov)
> Scale <- 3
> TwoDimEllipsoid(sample.mean, sample.cov, Scale) |
Projection Matrix has only 0 and 1 eigen values
> P <- A %*% solve(t(A) %*% A) %*% t(A)
> eigen(P)
$values
[1] 1 1 1
$vectors
[,1] [,2] [,3]
[1,] 0.7390028 -0.5345225 0.4100739
[2,] 0.2899661 0.8017837 0.5225539
[3,] -0.6081074 -0.2672612 0.7475138 |
Thus eigen vectors in the above matrix is 1 or 0.