Chi Plot
Purpose
Use Chi Plot to study randomness
The funda behind the plot is to look at a non parametric estimator of joint distribution of the 2 variables and marginals. Subsequently create a plot with respect to lambda versus chi.
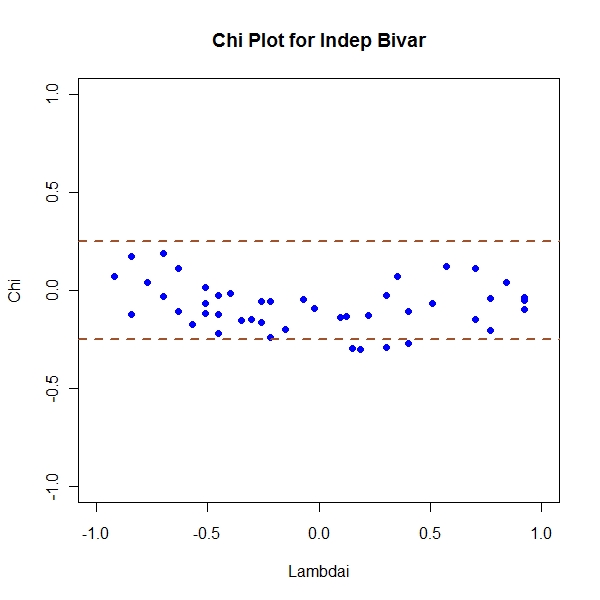
Independent Bivariates
> n <- 50 > x <- array(rnorm(50)) > y <- array(rnorm(50)) > z <- cbind(x, y) > Fi <- apply(x, 1, function(temp) length(which(x[which(x != temp)] <= + temp))/(n - 1)) > Gi <- apply(y, 1, function(temp) length(which(y[which(y != temp)] <= + temp))/(n - 1)) > Hi <- apply(z, 1, function(temp) length(which(x[which(x != temp[1])] <= + temp[1] & y[which(y != temp[2])] <= temp[2]))/(n - 1)) > Chi <- (Hi - Fi * Gi)/sqrt(Fi * (1 - Fi) * Gi * (1 - Gi)) > Lambdai <- 4 * sign((Fi - 0.5) * (Gi - 0.5)) * pmax((Fi - 0.5)^2, + (Gi - 0.5)^2) > result <- cbind(Lambdai, Chi) > condition <- abs(result[, 1]) < 4 * (1/(n - 1) - 0.5)^2 > not.consid <- n - sum(condition) > result <- (result[condition, ]) > cp <- 1.78 > par(mfrow = c(1, 1)) > plot(Lambdai, Chi, pch = 19, col = "blue", ylim = c(-1, 1), main = "Chi Plot for Indep Bivar") > abline(h = c(-1, 1) * cp/sqrt(n), col = "sienna", lwd = 2, lty = "dashed") |
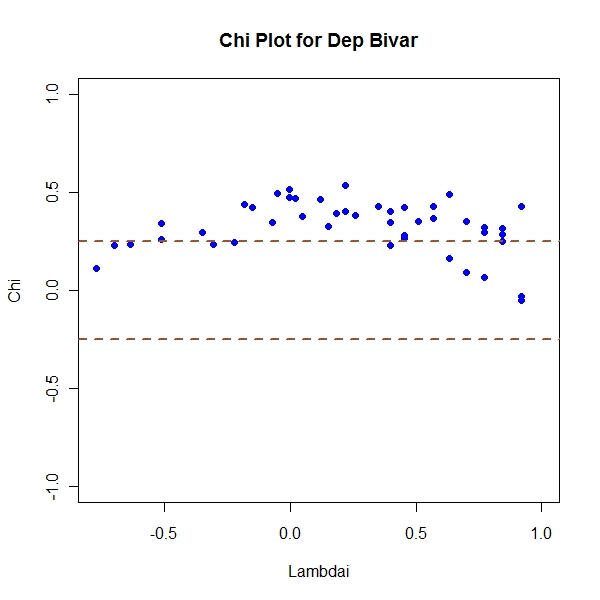
Dependent Bivariates
> library(mnormt) > n <- 50 > sample.mean <- c(0, 0) > sample.cov <- matrix(c(1, 0.7, 0.7, 1), nrow = 2) > z <- rmnorm(n, mean = sample.mean, varcov = sample.cov) > x <- array(z[, 1]) > y <- array(z[, 2]) > Fi <- apply(x, 1, function(temp) length(which(x[which(x != temp)] <= + temp))/(n - 1)) > Gi <- apply(y, 1, function(temp) length(which(y[which(y != temp)] <= + temp))/(n - 1)) > Hi <- apply(z, 1, function(temp) length(which(x[which(x != temp[1])] <= + temp[1] & y[which(y != temp[2])] <= temp[2]))/(n - 1)) > Chi <- (Hi - Fi * Gi)/sqrt(Fi * (1 - Fi) * Gi * (1 - Gi)) > Lambdai <- 4 * sign((Fi - 0.5) * (Gi - 0.5)) * pmax((Fi - 0.5)^2, + (Gi - 0.5)^2) > result <- cbind(Lambdai, Chi) > condition <- abs(result[, 1]) < 4 * (1/(n - 1) - 0.5)^2 > not.consid <- n - sum(condition) > result <- (result[condition, ]) > cp <- 1.78 > par(mfrow = c(1, 1)) > plot(Lambdai, Chi, pch = 19, col = "blue", ylim = c(-1, 1), main = "Chi Plot for Dep Bivar") > abline(h = c(-1, 1) * cp/sqrt(n), col = "sienna", lwd = 2, lty = "dashed") |
Let me see whether chiplot code can be made in to a function or not
> n <- 50
> sample.mean <- c(0, 0)
> sample.cov <- matrix(c(1, 0.7, 0.7, 1), nrow = 2)
> z <- rmnorm(n, mean = sample.mean, varcov = sample.cov)
> GetChiPlotData <- function(z) {
+ n <- dim(z)[1]
+ x <- array(z[, 1])
+ y <- array(z[, 2])
+ Fi <- apply(x, 1, function(temp) length(which(x[which(x !=
+ temp)] <= temp))/(n - 1))
+ Gi <- apply(y, 1, function(temp) length(which(y[which(y !=
+ temp)] <= temp))/(n - 1))
+ Hi <- apply(z, 1, function(temp) length(which(x[which(x !=
+ temp[1])] <= temp[1] & y[which(y != temp[2])] <= temp[2]))/(n -
+ 1))
+ Chi <- (Hi - Fi * Gi)/sqrt(Fi * (1 - Fi) * Gi * (1 - Gi))
+ Lambdai <- 4 * sign((Fi - 0.5) * (Gi - 0.5)) * pmax((Fi -
+ 0.5)^2, (Gi - 0.5)^2)
+ result <- cbind(Lambdai, Chi)
+ condition <- abs(result[, 1]) < 4 * (1/(n - 1) - 0.5)^2
+ not.consid <- n - sum(condition)
+ result <- (result[condition, ])
+ return(list(result = result, not.considered = not.consid))
+ }
> print(GetChiPlotData(z))
$result
Lambdai Chi
[1,] 0.57017909 0.3817040
[2,] -0.77009579 0.2211629
[3,] 0.40024990 0.3426714
[4,] 0.77009579 0.3185426
[5,] 0.84339858 0.3151982
[6,] -0.63348605 0.1819625
[7,] 0.15035402 0.6322076
[8,] 0.70012495 0.5172935
[9,] 0.63348605 0.4423879
[10,] 0.84339858 0.4787234
[11,] 0.18367347 0.4647580
[12,] 0.05039567 0.6446564
[13,] 0.35027072 0.3800585
[14,] -0.03373594 0.6475506
[15,] -0.22032486 0.5204165
[16,] 0.57017909 0.1964671
[17,] 0.40024990 0.5026842
[18,] 0.63348605 0.2233177
[19,] 0.35027072 0.4544504
[20,] 0.30362349 0.3271425
[21,] 0.51020408 0.1538968
[22,] 0.18367347 0.3302891
[23,] 0.57017909 0.1985712
[24,] -0.07038734 0.5176139
[25,] 0.22032486 0.4798488
[26,] -0.01041233 0.6023524
[27,] -0.12036651 0.5150819
[28,] 0.40024990 0.3567158
[29,] 0.30362349 0.1987769
[30,] -0.26030820 0.5579887
[31,] 0.51020408 0.2830693
[32,] 0.26030820 0.4455119
[33,] 0.77009579 0.1461864
[34,] 0.45356102 0.4024390
[35,] 0.84339858 0.2962532
[36,] -0.18367347 0.5252257
[37,] -0.45356102 0.3987333
[38,] 0.70012495 0.4714045
[39,] 0.51020408 0.4508348
[40,] -0.15035402 0.4624973
[41,] 0.57017909 0.4693807
[42,] 0.35027072 0.4557193
[43,] 0.35027072 0.3228829
[44,] 0.26030820 0.1831267
$not.considered
[1] 6 |
I will use this function from now on to see the dependency structure