Understanding Tail Dependencies using Chi-Plot
Purpose
Use Chi plots to look at 1. Tail Dependencies ? 2. Are tail dependencies symmetric or asymmetric ? 3. Spot Outliers
Prepare Data
> library(mnormt) > n <- 3000 > sample.mean <- c(0, 0) > sample.cov <- matrix(c(1, 0.7, 0.7, 1), nrow = 2) > z1 <- rmnorm(n, mean = sample.mean, varcov = sample.cov) > z2 <- rmt(n, mean = sample.mean, S = sample.cov, df = 3) > z1.x <- quantile(z1[, 1], prob = seq(0, 1, 0.05))[20] > z1.y <- quantile(z1[, 2], prob = seq(0, 1, 0.05))[20] > condition1 <- (z1[, 1] > z1.x) & (z1[, 2] > z1.y) > z2.x <- quantile(z2[, 1], prob = seq(0, 1, 0.05))[20] > z2.y <- quantile(z2[, 2], prob = seq(0, 1, 0.05))[20] > condition2 <- (z2[, 1] > z2.x) & (z2[, 2] > z2.y) > par(mfrow = c(2, 2)) > plot(z1, col = "blue", pch = 19, main = "Bivariate N", xlab = "X", + ylab = "Y") > plot(z2, col = "blue", pch = 19, main = "Bivariate t", xlab = "X", + ylab = "Y") > plot(z1[condition1, ], col = "blue", pch = 19, main = "Tails in Norm", + xlab = "X", ylab = "Y") > plot(z2[condition2, ], col = "blue", pch = 19, main = "Tails in t", + xlab = "X", ylab = "Y") |
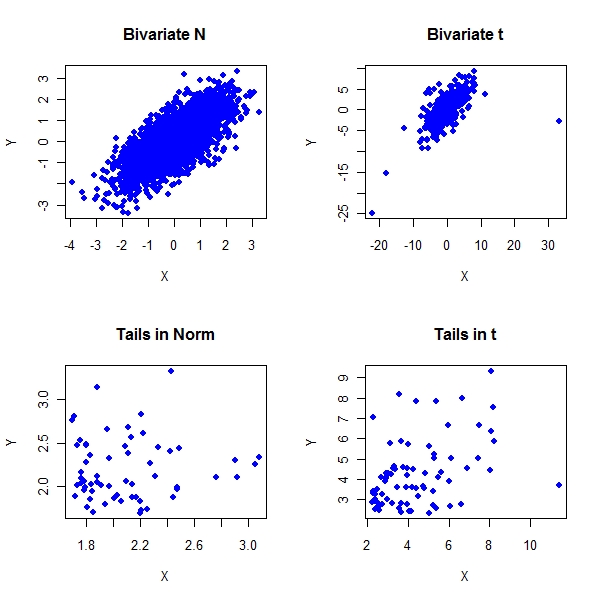
ChiPlots
> GetChiPlotData <- function(z) {
+ n <- dim(z)[1]
+ x <- array(z[, 1])
+ y <- array(z[, 2])
+ Fi <- apply(x, 1, function(temp) length(which(x[which(x !=
+ temp)] <= temp))/(n - 1))
+ Gi <- apply(y, 1, function(temp) length(which(y[which(y !=
+ temp)] <= temp))/(n - 1))
+ Hi <- apply(z, 1, function(temp) length(which(x[which(x !=
+ temp[1])] <= temp[1] & y[which(y != temp[2])] <= temp[2]))/(n -
+ 1))
+ Chi <- (Hi - Fi * Gi)/sqrt(Fi * (1 - Fi) * Gi * (1 - Gi))
+ Lambdai <- 4 * sign((Fi - 0.5) * (Gi - 0.5)) * pmax((Fi -
+ 0.5)^2, (Gi - 0.5)^2)
+ result <- cbind(Lambdai, Chi)
+ return(result)
+ }
> cp <- 1.78
> z1c <- GetChiPlotData(z1)
> z2c <- GetChiPlotData(z2)
> master1 <- cbind(z1, z1c)
> master2 <- cbind(z2, z2c) |
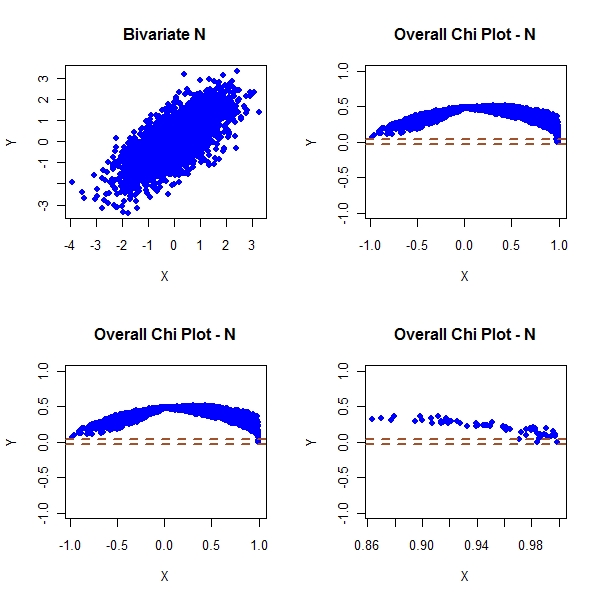
> par(mfrow = c(2, 2)) > plot(master1[, 1], master1[, 2], col = "blue", pch = 19, main = "Bivariate N", + xlab = "X", ylab = "Y") > plot(master1[, 3], master1[, 4], pch = 19, col = "blue", ylim = c(-1, + 1), main = "Overall Chi Plot - N", xlab = "X", ylab = "Y") > abline(h = c(-1, 1) * cp/sqrt(n), col = "sienna", lwd = 2, lty = "dashed") > plot(master1[!condition1, 3], master1[!condition1, 4], pch = 19, + col = "blue", ylim = c(-1, 1), main = "Overall Chi Plot - N", + xlab = "X", ylab = "Y") > abline(h = c(-1, 1) * cp/sqrt(n), col = "sienna", lwd = 2, lty = "dashed") > plot(master1[condition1, 3], master1[condition1, 4], pch = 19, + col = "blue", ylim = c(-1, 1), main = "Overall Chi Plot - N", + xlab = "X", ylab = "Y") > abline(h = c(-1, 1) * cp/sqrt(n), col = "sienna", lwd = 2, lty = "dashed") |
> par(mfrow = c(2, 2)) > plot(master2[, 1], master2[, 2], col = "blue", pch = 19, main = "Bivariate N", + xlab = "X", ylab = "Y") > plot(master2[, 3], master2[, 4], pch = 19, col = "blue", ylim = c(-1, + 1), main = "Overall Chi Plot - N", xlab = "X", ylab = "Y") > abline(h = c(-1, 1) * cp/sqrt(n), col = "sienna", lwd = 2, lty = "dashed") > plot(master2[!condition2, 3], master2[!condition2, 4], pch = 19, + col = "blue", ylim = c(-1, 1), main = "Overall Chi Plot - N", + xlab = "X", ylab = "Y") > abline(h = c(-1, 1) * cp/sqrt(n), col = "sienna", lwd = 2, lty = "dashed") > plot(master2[condition2, 3], master2[condition2, 4], pch = 19, + col = "blue", ylim = c(-1, 1), main = "Overall Chi Plot - N", + xlab = "X", ylab = "Y") > abline(h = c(-1, 1) * cp/sqrt(n), col = "sienna", lwd = 2, lty = "dashed") |
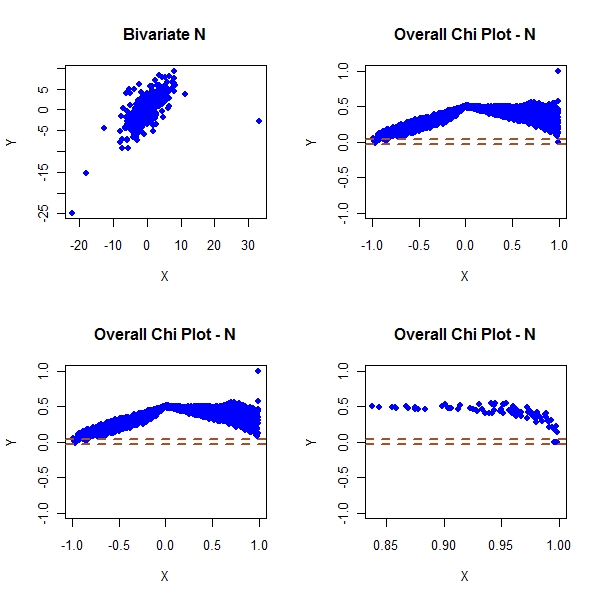
Look at the above plots. You can clearly see the tail dependence
The t joint dist shows that the tail dependence no longer goes to 0. This is unlike gaus joint which shows that tail dependence goes to 0.
A marvelous way to visualize tail dependencies.