Dobson - GLM - Chap 1
Purpose
Exercise problems in Chap 1
The data below are the numbers off emales and males in the progeny of 16 female light brown apple moths in Muswellbrook, New South Wales, Australia (from Lewis, 1987).
Find the maximum likelihood estimator of theta using calculus and evaluate it for these data if each of Y’s are independent binomial realizations .Here’s the data
> x <- read.csv("test.csv", header = T, stringsAsFactors = F)
> x$total <- x$f + x$m
> print(x)
group f m total
1 1 18 11 29
2 2 31 22 53
3 3 34 27 61
4 4 33 29 62
5 5 27 24 51
6 6 33 29 62
7 7 28 25 53
8 8 23 26 49
9 9 33 38 71
10 10 12 14 26
11 11 19 23 42
12 12 25 31 56
13 13 14 20 34
14 14 4 6 10
15 15 22 34 56
16 16 7 12 19 |
Coding up the likelihood function
> loglikf <- function(theta, z) {
+ lik <- 0
+ for (i in seq_along(x[, 1])) {
+ t1 <- choose(x$total[i], x$f[i])
+ t2 <- theta^(x$f[i])
+ t3 <- (1 - theta)^(x$m[i])
+ temp <- t1 * t2 * t3
+ lik <- lik + temp
+ }
+ return(log(lik))
+ } |
Using basic graph
> thetas <- seq(0.01, 0.99, 0.001)
> result <- numeric(0)
> for (i in seq_along(thetas)) {
+ result[i] <- loglikf(thetas[i], z)
+ }
> plot(thetas, result, pch = 19, col = "blue", xlab = "theta",
+ ylab = "loglik", main = "") |
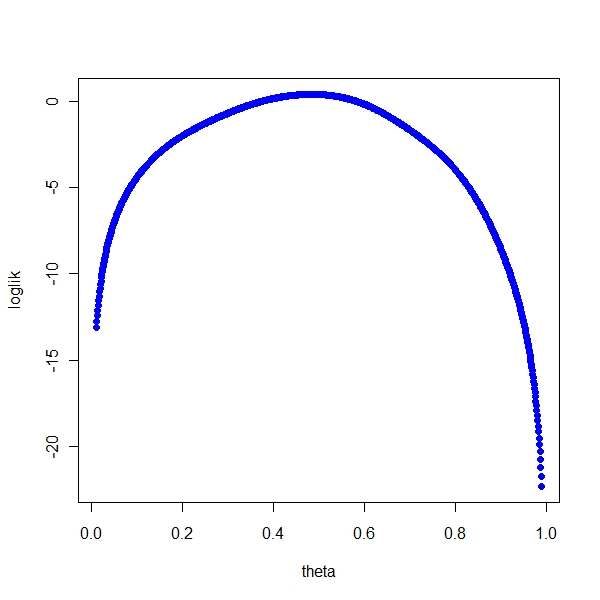
Using optim
> theta0 <- 0.1 > solution <- optim(par = theta0, fn = loglikf, control = list(z = x, + fnscale = -2)) > print(solution$par) [1] 0.4838281 |
Reconciling both
> print(solution$par) [1] 0.4838281 > thetas[which.max(result)] [1] 0.484 |